RESEARCH
Transposable elements, genome stability & evolution
A large fraction of our genome (up to 50%) consists of selfish DNA modules known as transposable elements (TEs) – mobile units that aim to increase in copy number by jumping from one location to the other. Since their discovery, transposable elements have been involved in the organization, functioning, and evolution of genomes, but their uncontrolled activity can be detrimental to the host and must therefore be regulated. This is especially true in the germline, the battleground where host and transposon mechanisms compete for
maximizing their influence over the genetic
information that will be passed down to the next
generation. On one side, selfish elements aim to
increase in copy number by mobilization (Jansen et
al, 2024), while on the other side host mechanisms
act to suppress TE activity and its detrimental effects
(Teixeira et al, 2017). Often balanced, these two
opposed actions are thought to safeguard the
functional and structural integrity of the genome
while allowing genome evolution (Gebert et al, 2021;
Gebert et al, 2025). Together with Richard Durbin's
group, we are building an interdisciplinary research team that will combine large-scale experimental evolution studies and innovative computational methods to uncover the breadth of transposon-derived genetic variation and its role in genome evolution in invertebrate and vertebrate systems.
In parallel, we have been taking advantage of a classic example of disruptive transposon activity provided by P-M hybrid dysgenesis in Drosophila to study how transposons are controlled and how the germline copes with TE activity once they become active. P-M dysgenesis is a syndrome that specifically affects germline development and is triggered by the P-element DNA transposon (Malone et al, 2015). In this system, the presence of epigenetically inherited small RNAs (sRNAs) complementary to the P-element correlates with germline protection. Unexpectedly, we found that such sRNAs regulate P-element splicing, leading to the accumulation of inactive transcripts in Drosophila germ cells (Teixeira et al, 2017). Using this textbook example of genomic conflict and transgenerational epigenetics, we use a combination of developmental and high-throughput molecular approaches to dissect the host mechanisms controlling genome stability and transposon activity during germline development (Jansen et al, 2024).

Regulation of gene expression during germline development
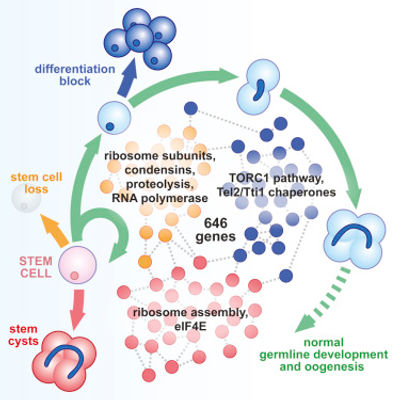
Germ cells are considered to be the ultimate stem cell lineage because they have the ability to generate more germ cells as well as give rise to a new organism for every generation. Not surprisingly, the study of the germline has been instrumental in building the understanding of the mechanisms of pluripotency and stem cell biology, with broad implications for regenerative medicine, ageing, and cancer. We have previously assembled the complete genetic framework controlling germline stem cell self-renewal and differentiation in vivo (Sanchez et al, 2016). This was accomplished by performing an unbiased,
transcriptome-wide in vivo RNAi screen, followed
by in-depth developmental, genetic, and
computational analyses. Further analyses led us
to discover exciting new aspects of germline
development, including the translational
control of stem cell cytokinesis, the uncoupling of
ribosome biogenesis and protein synthesis rates
during stem cell development, and an energy
metabolism-independent function for mitochondria
governing differentiation (Teixeira et al, 2015;
Sanchez et al, 2016; Gui et al, 2023; Samuels et al,
From plants to mammals, accumulated evidence indicate that regulation of protein synthesis plays critical roles during organism development, tissue homeostasis, and tumorigenesis (Teixeira & Lehmann, 2019). However, the study of spatiotemporal regulation of gene expression and its function in controlling stem cell biology has been mostly restricted to chromatin-based mechanisms. Interestingly, translational regulation appears prominent in the germline, as no master transcription factor has been identified to specify germ cell fate. Using a combination of high-throughput molecular approaches, genomics, as well as developmental and genetic analyses, we are dissecting the molecular mechanisms by which protein synthesis control governs key aspects of germline stem cell development in vivo (Gui et al, 2023; Samuels et al, 2024).